Mass spectrometry-based methods to study protein amyloid formation
An increasing number of proteins are known to form amyloid fibrils in vivo, and the formation of these fibrils is implicated in several diseases (e.g. Alzheimer's, Parkinson's). One of these proteins, β-2-microglobulin (β2m), can form amyloid fibrils, and these fibrils are the main pathogenic process underlying dialysis-related amyloidosis (DRA). Like other amyloid systems, β2m fibril formation proceeds by partial protein unfolding, subsequent oligomerization, and eventual elongation to form mature fibrils. While aspects of general amyloid formation are understood, molecular-level information about the early stages of the amyloid reaction is lacking for almost all amyloid systems; however, this information is critical for the rational design of therapeutics against amyloid diseases. We strive to obtain amino acid-level information of the unfolding and oligomerization of β2m prior to fibril formation by developing mass spectrometry-based methods with the necessary temporal and spatial resolution. We are particularly interested in the role that Cu(II) plays in the amyloid formation of β2m. In general, the methods that we develop not only provide insight into β2m's amyloidosis but are also general enough to work for other amyloid systems.
Nanomaterials as novel extraction/concentration/detection methods for protein analyses in complex mixtures.
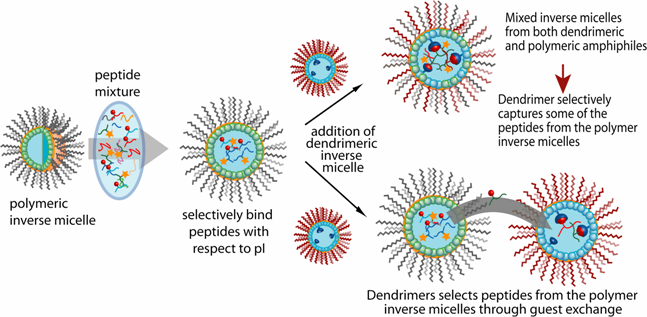
Nanomaterials have great potential for enhancing protein analysis in complex mixtures. We use functionalized nanomaterials as new extraction/concentration/detection methods that can more fully exploit the exquisite sensitivity and specificity of mass spectrometry. The "bottom-up" design control of these materials from the molecular level to the micron/millimeter level makes them adaptable tools for coupling with mass spectrometry. Together with mass spectrometry, these nanomaterials are being used in several contexts: (1) to extract proteins and peptides in complex mixtures with high selectivity and efficiency; (2) to fractionate protein and peptide mixtures according to desired chemical or physical properties; (3) to facilitate protein identification in complex mixtures; and (4) to detect compounds of interest in cell lysates and whole cells.
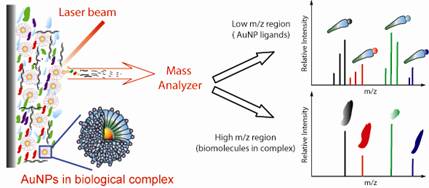 |
We are also developing new mass spectrometry-based approaches to track, measure, and image nanoparticles in complex samples. These methods have two particular applications. First, nanoparticles are increasingly being investigated as therapeutic delivery agents, and effective development of such delivery agents requires the ability to monitor nanoparticles in cells, tissues and/or organs. Second, nanomaterials are more and more found in commercial products, and their widespread use is starting to raise concerns about their impact on the environment. New measurement tools are therefore needed to understand the fate, transport, and bioavailability of such engineered nanoparticles
Protein Therapeutics
Protein therapeutics account for about one-third of the late-stage drug development pipeline. The higher order structure (HOS) of these therapeutics is essential to monitor to ensure their efficacy and safety. HOS changes can lead to reduced stability, loss of efficacy, and possible immunogenicity. We have explored the use of covalent labeling and mass spectrometry (CL-MS) as a means to detect HOS changes to therapeutic proteins, with human growth hormone and immunoglobulin IgG1 as model systems. Diethylpyrocarbonate (DEPC) CL-MS provides moderate structural resolution in a relatively efficient manner. Currently we are investigating the merits of our DEPC CL-MS technology to detect subtle conformation changes to therapeutic antibodies.
