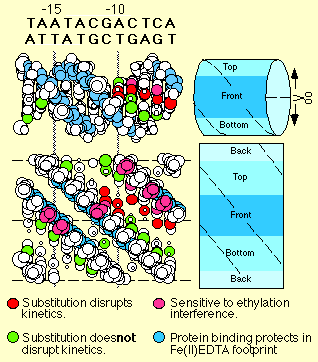
Another research focus of the group is the study of the enzymology of transcription. Specifically, we are currently probing structural details of the protein-DNA interface in the recognition of the promoter sequence by T7 RNA polymerase. In this work, we have used "functional group mutagenesis," that is the chemical synthesis of DNA in which a single functional group within the promoter sequence is altered, providing a very simple probe of important contacts. Results from studies of the upstream part of the promoter are summarized in the figure below, in a GIF animation, or in a CHIME Window (you must have the CHIME plug-in installed).

The upper part of the figure shows a space filling model of a portion of the promoter DNA, while the lower figure shows the same DNA in an expanded form which allows all sides of the DNA to be viewed at once. The functional groups identified in binding (red) all lie along one face of the predicted DNA duplex, while groups not involved in binding (green) lie along the opposite side.
These data strongly support a model in which the enzyme directly "reads out" the sequence of the DNA by contacting specific amino, carboxyl, and methyl groups within the central major groove of the DNA.
Although this simple model is supported in the upstream part of the promoter sequence (9 to 12 bases from the start site for transcription), a variety of data now indicate that nearer the start site (position +1) the enzyme interacts only with one strand of the DNA. The "template strand" ultimately serves as a template for the sequence specific synthesis of the resulting RNA.
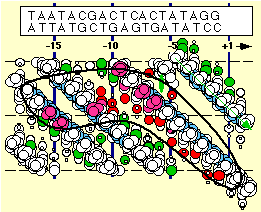
Date for the entire promoter are summarized above, and indicate clearly the recognition of duplex DNA upstream and the recognition of only the template strand downstream.
Future studies of this enzyme will focus on obtaining a more detailed understanding of the kinetic steps in the synthesis of a very short (5 base) RNA polymer. Using a rapid quench flow instrument, we can initiate a reaction, and then stop, or "quench," the reaction within a reaction time as short as 10 msec. Final and intermediate products can then be followed as a function of time.
These kinds of experiments will be extremely valuable in unraveling the mechanism of the early stages of transcription. We will ultimately combine this approach with functional group mutagenesis in order to understand the mechanistic roles of the structural contacts described above.
T7 RNA polymerase, whose structure (PDB file 2RNP) has been determined (in the absence of DNA), and which is structurally homologous to a family of DNA and RNA polymerases (alignments), presents an ideal model system in which to study fundamental aspects of transcription.